
(BLOOMINGTON) – Indiana University microbiologist Malcolm Winkler has been awarded a $3.3-million, five-year grant from the NIH National Institute of General Medical Sciences to study the dynamics and regulation of peptidoglycan (PG) cell wall synthesis in the “superbug” Streptococcus pneumoniae (pneumococcus).
Dr. Winkler is a professor in the Department of Biology and adjunct core professor in the Department of Molecular and Cellular Biochemistry in the IU Bloomington College of Arts and Sciences. The grant is an Established Investigator MIRA (Maximizing Investigators’ Research Award) that consolidates and renews previous NIH RO1 and Multiple Principal Investigator (MPI) grants to the Winkler laboratory.
Antibiotic resistance and evasion of treatment are increasing at alarming rates for numerous important bacterial pathogens—including S. pneumoniae—and represent a serious threat to human health worldwide. S. pneumoniae alone causes about 4 million infections (1.2 million that are drug-resistant) and about 25,000 deaths per year in the U.S.A. and over 1 million deaths per year worldwide, according to Dr. Winkler.

The Winkler lab uses the “superbug” respiratory pathogen S. pneumoniae as a highly tractable model for ovoid-shaped pathogenic bacteria to understand different levels of regulation of PG cell wall synthesis. New findings about regulatory interactions and circuits have the potential to provide targets and reveal new vulnerabilities for future vaccine and antibiotic discovery.
The PG cell wall is a gigantic mesh-like polymer that protects bacteria from osmotic damage and determines cell size, shape, and chaining—required for survival in human hosts and environmental niches. In Gram-positive bacteria like S. pneumoniae, PG also acts as the scaffold for covalent attachment of other surface macromolecules required for growth, adhesion, and virulence interactions. The regulation of PG synthesis is a fundamentally important spatial and temporal biological problem that involves interactions, assembly, and disassembly of a large ensemble of proteins and expression of these proteins at levels that are correct for normal growth and changed during stress.
In S. pneumoniae cells, the midcell equator of newly divided cells is the site of future cell division and septum closure by PG synthesis (see figure). The midcell is also the site for side-wall-like PG elongation (peripheral synthesis) that gives S. pneumoniae cells their characteristic American-football shape. PG synthesis is carried out by enzyme complexes that contain penicillin-binding proteins (PBPs), which are the targets of β-lactam antibiotics such as penicillin. PBPs interact with other PG synthases and regulatory proteins, while PG hydrolases cut and remodel the PG mesh structure. The molecular nano-machines that carry out septal and peripheral PG synthesis are organized at equatorial midcells by the dynamics of tubulin-, actin-, and spectrin-like bundles and filaments.
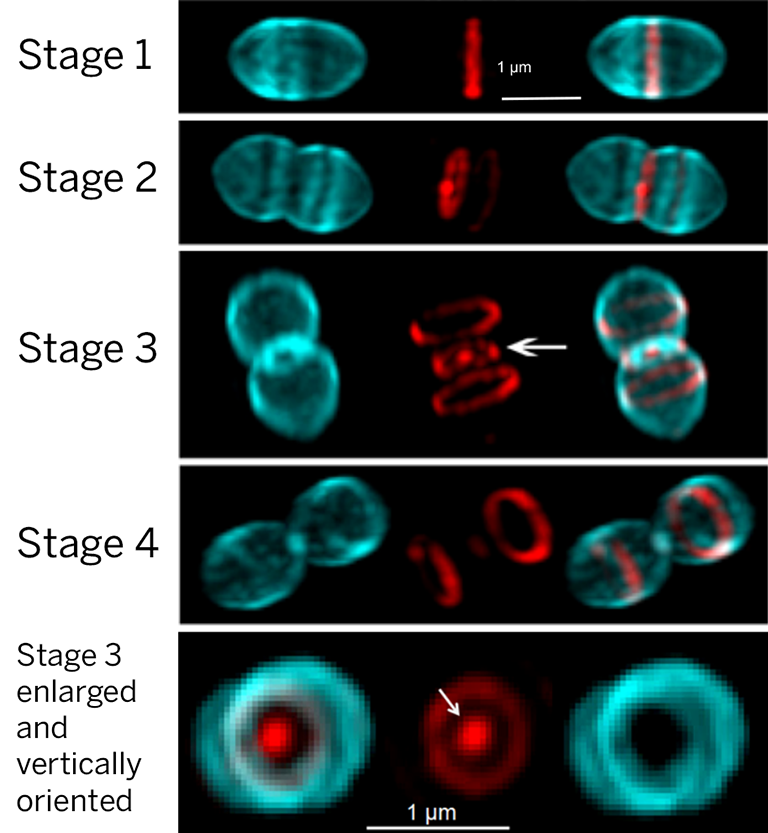
The MIRA grant will allow the Winkler lab to determine the composition, chronology, coordination, and regulation of these protein molecular machines at different stages of pneumococcal cell division. It will also support the lab in explicating mechanisms that control amounts of precursors required for PG synthesis. Questions about PG synthesis dynamics and regulation are being answered with a powerful combination of molecular genetic, physiological, cell biological (including high-resolution, TIRF, and single-molecule microscopy), and biochemical methods (including MS-based proteomics and UHPLC-MS determinations) to attack this multicomponent problem. Together, these findings will fill in major knowledge gaps about the regulation of PG synthesis in a model ovoid-shaped bacterium, identify functions of reported virulence factors, and provide new extracellular targets for antibiotics and vaccines.
The project has benefited greatly from collaborations with the IU Bloomington College of Arts and Sciences laboratories of Professors David Giedroc and Michael VanNieuwenhze (Chemistry) and Sidney Shaw (Biology), as well as from the outstanding light microscopy, biological mass spectroscopy, and genomics and bioinformatics cores on campus.
Information, Indiana University, https://biology.indiana.edu



